Yaohai Bio-Pharma prepares circRNA based on the PIE system (alignment of exons and introns), which relies on the self-splicing function of type I introns to achieve RNA cyclization. The PIE structure is designed using the T4 td gene or fishy tRNA precursor gene, and the arrangement is as follows:
The RNA intron and supporting exon fragment are divided into two parts (5' terminal and 3' terminal), where the 5' terminal sequence is transferred to the tail of the target sequence, the 3 ' terminal sequence is inserted into the front of the target sequence, and the target gene sequence is inserted in the middle.
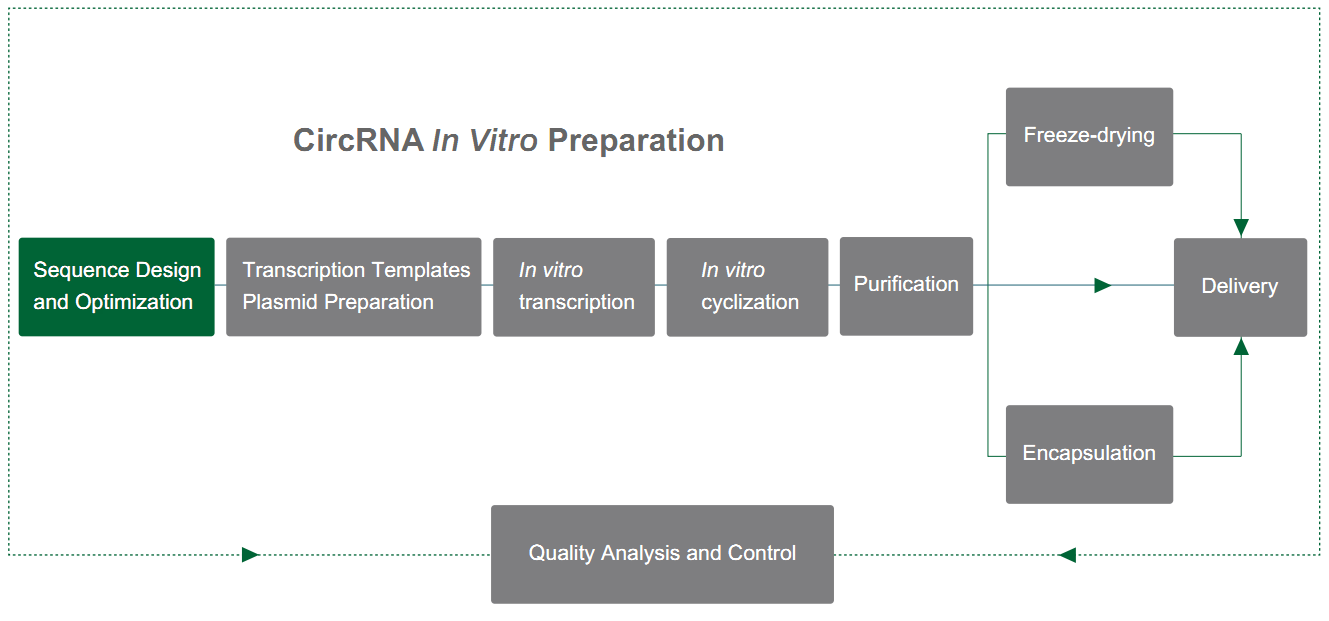
Under the catalysis of GTP, the PIE structure leads to the cyclization of sequences other than introns. Combined with a reasonable enhancement strategy of cyclization rate, Yaohai Bio-Pharma can achieve cyclization of sequences up to 4 kb with a cyclization rate of more than 80%.
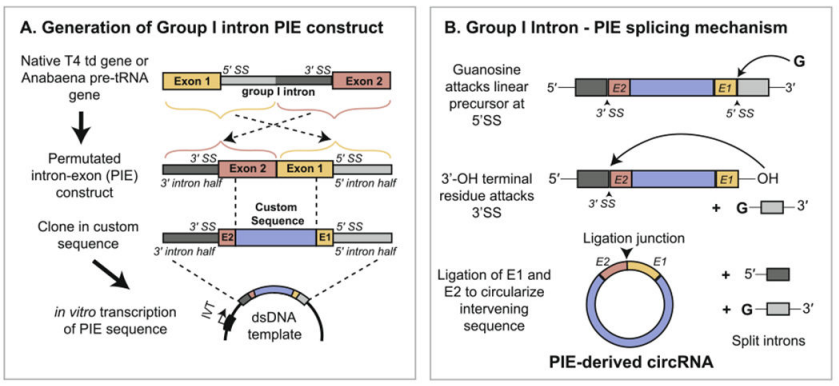
Figure 1. In vitro circulation of circRNA based on the PIE system
Services Details
| Process |
Optional Service |
Service Details |
Delivery Period (workday) |
| circRNA sequencedesign and optimization |
Design and optimization of coding sequences |
CDS sequence alignment
CDS codon optimization
|
1 |
| Design and optimization of non-coding sequences |
Intron and exon sequence design and optimization
Homologous arm sequence design and optimization
Spacer sequence design and optimization
|
1-2 |
Common Strategies for mRNA Sequence Design
| CircRNA Components |
Biological Functions |
Optimization Strategies |
| Two-terminal intron and exon sequences |
GTP-catalyzed intron self-splicing for cyclization of sequences outside the intron. |
Designed according to the T4 td gene or fish oil tRNA precursor gene. |
| Coding |
IRES |
Internal ribosome recognition site that regulates the translation of circRNA. |
Screening of internal ribosome entry site (IRES) sequences from different viral sources, e.g. EMCV, CVB3 sources. |
| CDS |
Protein-coding regions, sequences coding for antigens, antibodies or other functional proteins. |
Codon optimization increases the level of translation; certain non-optimal codons may play a role in protein folding. |
| Non-coding |
Non-coding sequences |
Target miRNAs or proteins to exert gene or protein regulation. |
Targeting specific binding sites for miRNAs or proteins can repeat the sequences of the binding site. |
Our Features
- Optimized PIE Cyclization System Combination with reasonable optimization strategies to achieve a cyclization rate of more than 80%;
- Cutting-edge CDS Optimization Team Cooperation with professional AI algorithm team to complete the optimization of CDS region codons;
- Mature and Perfect Process circRNA can be achieved with high cyclization efficiency, high stability and high translation efficiency.
Case study
Yaohai Bio-Pharma launched the control product enhanced green fluorescent protein (eGFP) circRNA, which is based on the PIE system to achieve the cyclization of RNA.
Using a conventional transfection reagent, eGFP circRNA is transfected into 293T cells, and eGFP (green) fluorescent signal can be detected after 24h, which will be enhanced after 48h.The fluorescent signal can still be detected on the 7th day and the 14th day after transfection.
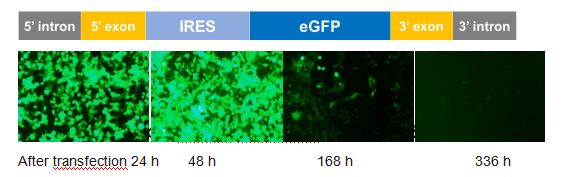
In vitro expression validation of eGFP circRNA

 EN
EN
 AR
AR
 HR
HR
 CS
CS
 DA
DA
 NL
NL
 FI
FI
 FR
FR
 DE
DE
 EL
EL
 IT
IT
 JA
JA
 KO
KO
 NO
NO
 PL
PL
 PT
PT
 RO
RO
 RU
RU
 ES
ES
 SV
SV
 IW
IW
 ID
ID
 LV
LV
 LT
LT
 SR
SR
 SK
SK
 SL
SL
 UK
UK
 VI
VI
 ET
ET
 HU
HU
 TH
TH
 TR
TR
 FA
FA
 AF
AF
 MS
MS
 BE
BE
 MK
MK
 UR
UR
 BN
BN