mRNA 3’ poly(A) Characterization
mRNA 3’ poly(A) Characterization
The 3’ poly(A) tail (polyadenosines) of mRNA is integral to post-transcriptional regulation, influencing mRNA export, stability, and translation. In the case of in vitro-transcribed (IVT) mRNAs, the poly(A) tail is introduced either during co-transcription in a templated manner or through an enzymatic tailing process in a non-templated manner. This 3′ poly(A) serves as a critical quality attribute (CQA) for IVT RNAs, necessitating thorough characterization during both process development and batch release.
Yaohai Bio-Pharma provides specialized 3’ poly(A) characterization services, employing techniques such as ribonuclease T1 (RNase T1) cleavage, Ion-pair reversed-phase high performance liquid chromatography (IP-RP-HPLC)/liquid chromatography-mass spectrometry (LC-MS), and comprehensive data analysis.
Regulatory Requirements for mRNA 3’ poly(A) Characterization
According to the WHO regulatory considerations, “the integrity of the structure of the mRNA is considered to be a critical quality attribute for the release of the mRNA. Thus, control is needed for mRNA integrity, 5′ capping efficiency, 3′ poly(A) tail presence or length and so on.”
Analytical Method
Analysis | Methods |
mRNA 3' poly(A) characterization | LC-MS after digestion |
Procedure
Step 1. RNase T1 cleavage
Direct analysis of full-length intact mRNA by LC-MS is impractical due to its size and heterogeneity. To examine the poly(A) tail, it must be cleaved from the full-length mRNA. The prevalent method involves enzymatic digestion with RNase T1, selectively cleaving the 3′ end of rG while preserving the poly(A) sequence. The mRNA sample undergoes digestion with RNase T1, and cleavage fragments containing poly(A) stretches are isolated using oligo dT-coated magnetic beads.
Step 2. IP-RP-HPLC/LC-MS
The extracted tails underwent analysis using LC-MS, offering precise tail length information at single-nucleotide resolution. In oligonucleotide LC-MS analysis, ion-pairing reversed-phase (IP-RP) separations with amine mobile phase additives stand as the preferred chromatographic mode, owing to their robust resolving power and compatibility with mass spectrometry (MS).
Step 3. Data analysis
The poly(A) tail length and its relative abundance depend on the number and intensity of peaks divided by a mass of adenosine. Upon processing or deconvoluting the mass spectrum, the anticipated mass of a 100-mer poly(A) tail (33,163 Da in box) is discerned, accompanied by a series of masses spaced by 329 amu, equivalent to the mass of adenosine (A).
Case of 3’ poly(A) Characterization with LC–MS
Yaohai-BioPharma has developed a robust method for mRNA 3’ poly(A) characterization, including RNase T1 cleavage and IP-RP-HPLC/LC-MS.
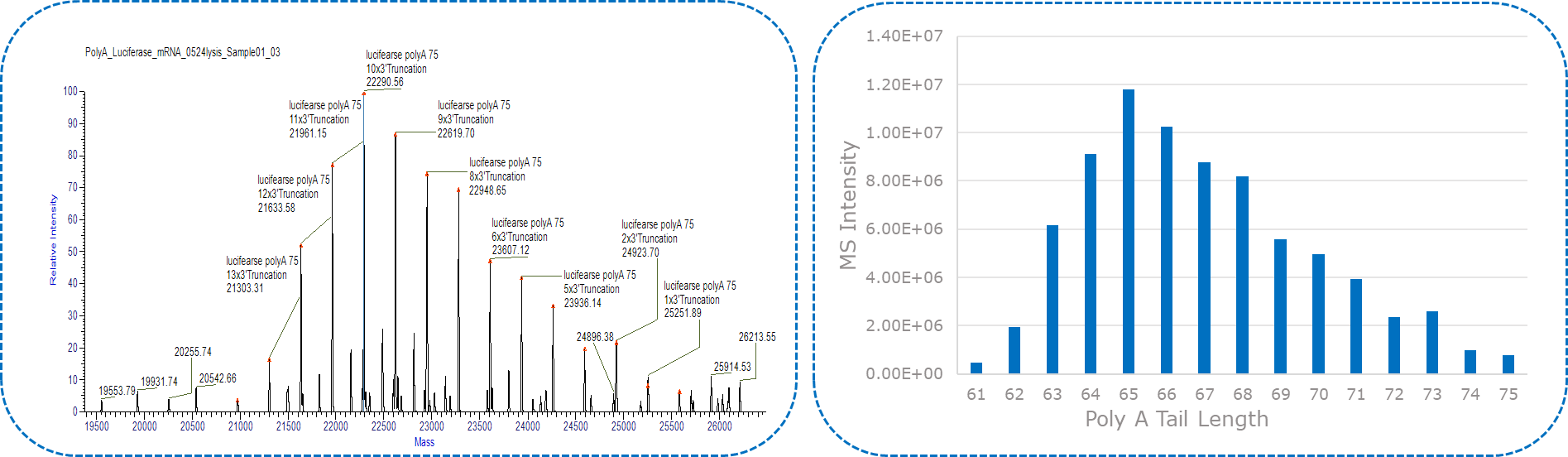
Yaohai-BioPharma has developed a robust method for mRNA 3’ poly(A) characterization, including RNase T1 cleavage and IP-RP-HPLC/LC-MS.

 EN
EN



